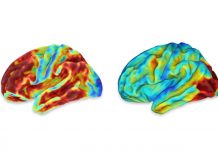
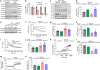
GAIT-GM Annotation Tool effectively maps genes and metabolites to pathways
Annotation using GAIT-GM text mining (see Supplementary Methods) resulted in 110/110 metabolites (100%) mapping to KEGG compound IDs and 3096/3146 genes (98%) mapping to KEGG IDs. While some gene and metabolite KEGG IDs did not have associated pathways, 79 metabolites were mapped to a total of 75 unique pathways, and 1187 genes mapped to a total of 115 unique pathways (Fig. S1). Pathways where both metabolites and genes were found included valine, leucine and isoleucine degradation; alanine, aspartate and glutamate metabolism; glycolysis / gluconeogenesis; and citrate cycle (TCA cycle), which recapitulates the pathway identification obtained at the separate analysis of these data6.
GAIT-GM Integrative Tools characterize the gene-metabolite co-variation network
We first hypothesized that metabolic changes in…