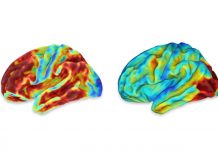
Manifold learning distinguishes pathologically defined LOAD from control
We first quantify the bulk RNA-Seq data from the ROS/MAP and Mayo Clinic cohorts into gene counts and remove any batch effects introduced due to sequencing runs using standard count normalization (see “Methods”). The data from the ROS/MAP cohort are sampled from the dorsolateral prefrontal cortex (DLPFC), and the data from the Mayo Clinic cohort are sampled from the temporal cortex (TCX). Patient’s clinical characteristics are reported in Supplementary Table 1 and described in “Methods.” The full pipeline we used for RNA-Seq data generation and quality control was recently reported22. The entire transcriptome comprises many genes, which do not have measurable expression or vary across case/control samples, which we remove in order to reduce the noise in manifold learning19. To do this, we first perform…